Epigenomic Regulation of Schwann Cell Reprogramming in Peripheral Nerve Injury | Journal of Neuroscience

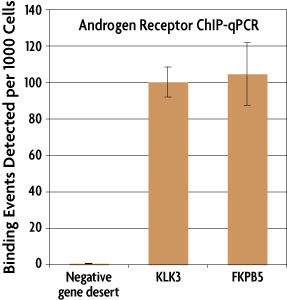
A laboratory practical illustrating the use of the ChIP‐qPCR method in a robust model: Estrogen receptor alpha immunoprecipitation using Mcf‐7 culture cells - Lacazette - 2017 - Biochemistry and Molecular Biology Education -
Epigenetic Landscape of Kaposi's Sarcoma-Associated Herpesvirus Genome in Classic Kaposi's Sarcoma Tissues | PLOS Pathogens

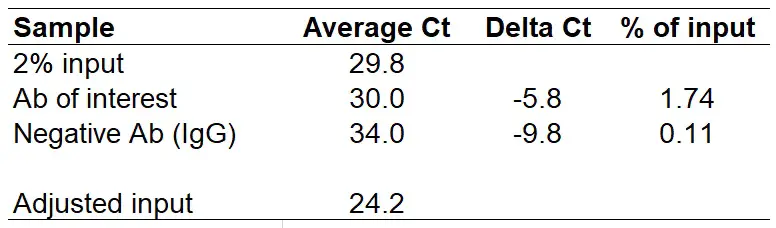
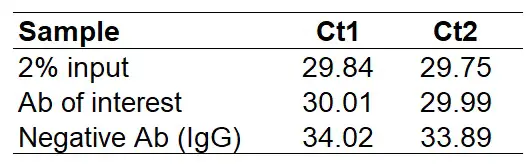
Example of ChIP-qPCR calculations and data presentation. (a) ChIP-qPCR... | Download Scientific Diagram
Unexpected binding behaviors of bacterial Argonautes in human cells cast doubts on their use as targetable gene regulators | PLOS ONE

Protocol for using heterologous spike-ins to normalize for technical variation in chromatin immunoprecipitation
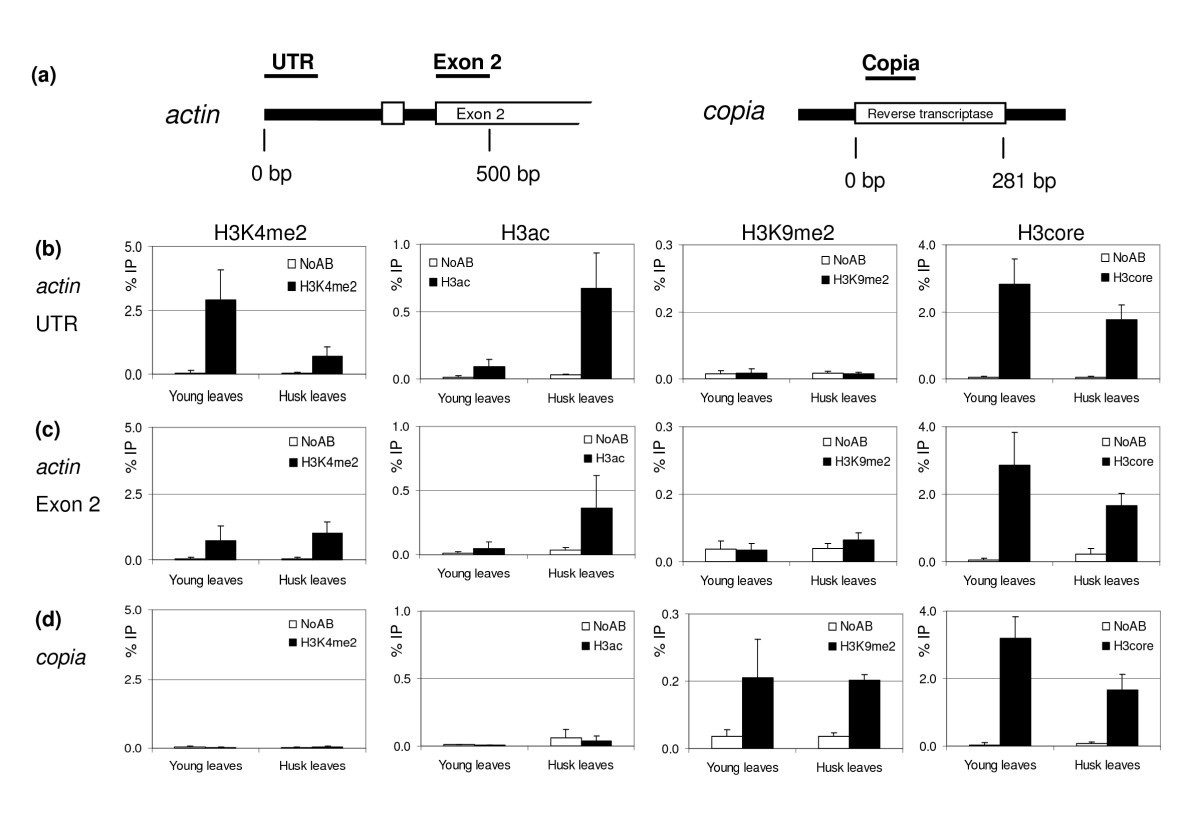
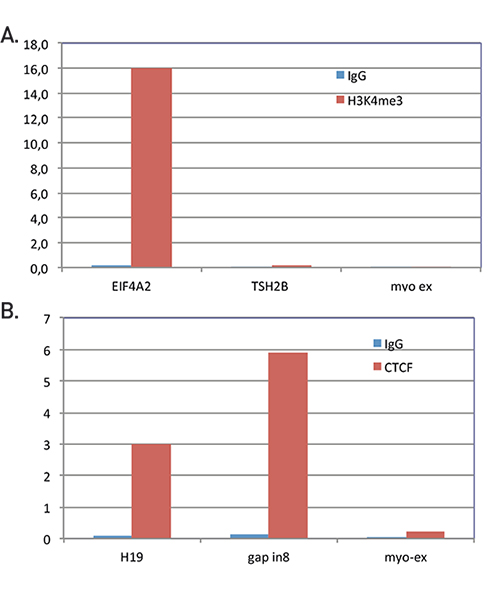
Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization | Plant Methods | Full Text

A laboratory practical illustrating the use of the ChIP‐qPCR method in a robust model: Estrogen receptor alpha immunoprecipitation using Mcf‐7 culture cells - Lacazette - 2017 - Biochemistry and Molecular Biology Education -